Dr. Kirsten Miller-Duys
29/07/2021
500 Views
![]()
At Hyrax Biosciences we continue to support the surveillance efforts for SARS-COV-2, to be able to detect and monitor Variants of Concern (VoC) and Variants of Interest (VoI). Our software platform Exatype makes the data analysis for next generation sequencing (NGS) data a simple, automated step in the genotyping process.
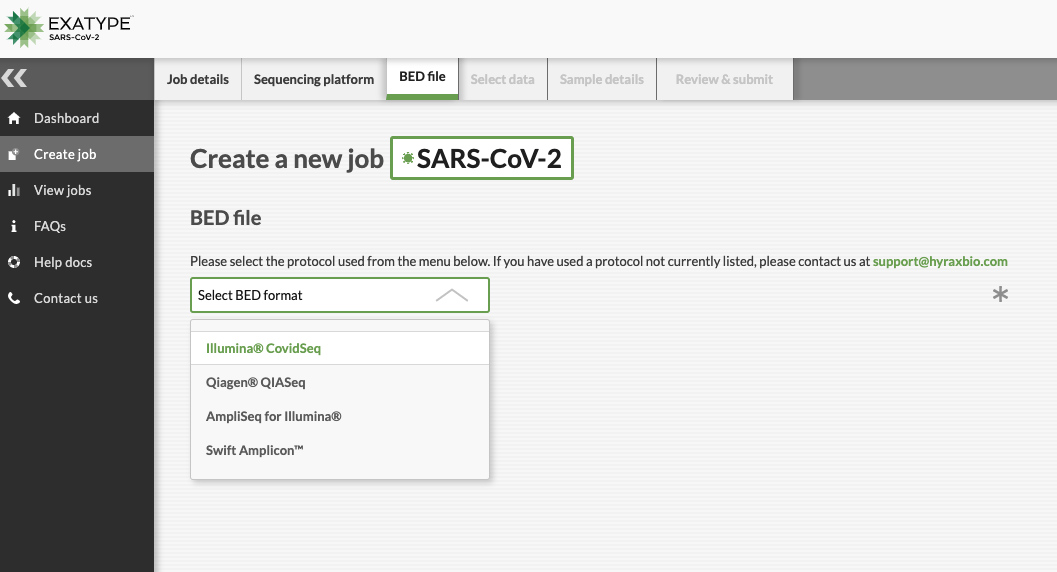
We have now expanded the type of analysis options available on Exatype. We continue to support the ARTIC and CleanPlex (from Paragon Genomics) protocols with the addition of 4 other protocols. You can now select from the following options:
- ARTIC (v1;v2;v3)
- CleanPlex® SARS-CoV-2 Panel (Flex and Flexv2)
- Illumina® CovidSeq
- AmpliSeq for Illumina®
- Qiagen® QIASeq
- Swift S-gene™
The intuitive front-end allows you to upload your sequence data files, then simply select the relevant protocol from our dropdown list.
Exatype can produce a consensus sequence within minutes. We use the latest versions of both Nextstrain (clade typing) and Pangolin (lineage typing) for reporting, so that you don’t need to worry about updating software.
For more in-depth information view our flyer or sign up for an account and take advantage of 10 free samples to see how Exatype can work for you!
If you would like Hyrax to support your company’s sample prep and sequencing workflow, get in touch today to see how we can work together to add your protocol to our DNA analysis platform.
 Your sequence data.
Your sequence data.