Concerns around the emergence of new SARS-CoV-2 variants that have increased transmissibility and/or reduce vaccine efficacy means that countries are continuing to scale their SARS-CoV-2 genomic surveillance.
There are two main reasons why genomic surveillance is performed:
- To monitor what variants are dominating infections within a country.
For example, in South Africa, the beta variant dominated early infections with the delta variant emerging from June 2021 onwards to currently comprise 100% of sequenced viruses (Figure 1). Such monitoring is critical in order to direct the public health response - the delta variant is thought to be as much as 60% more transmissible than previous variants and, thus, upon identification of the emergence of the delta variant, South Africa entered a strict lockdown in June 2021 to curtail transmission and reduce the impact of the third wave.
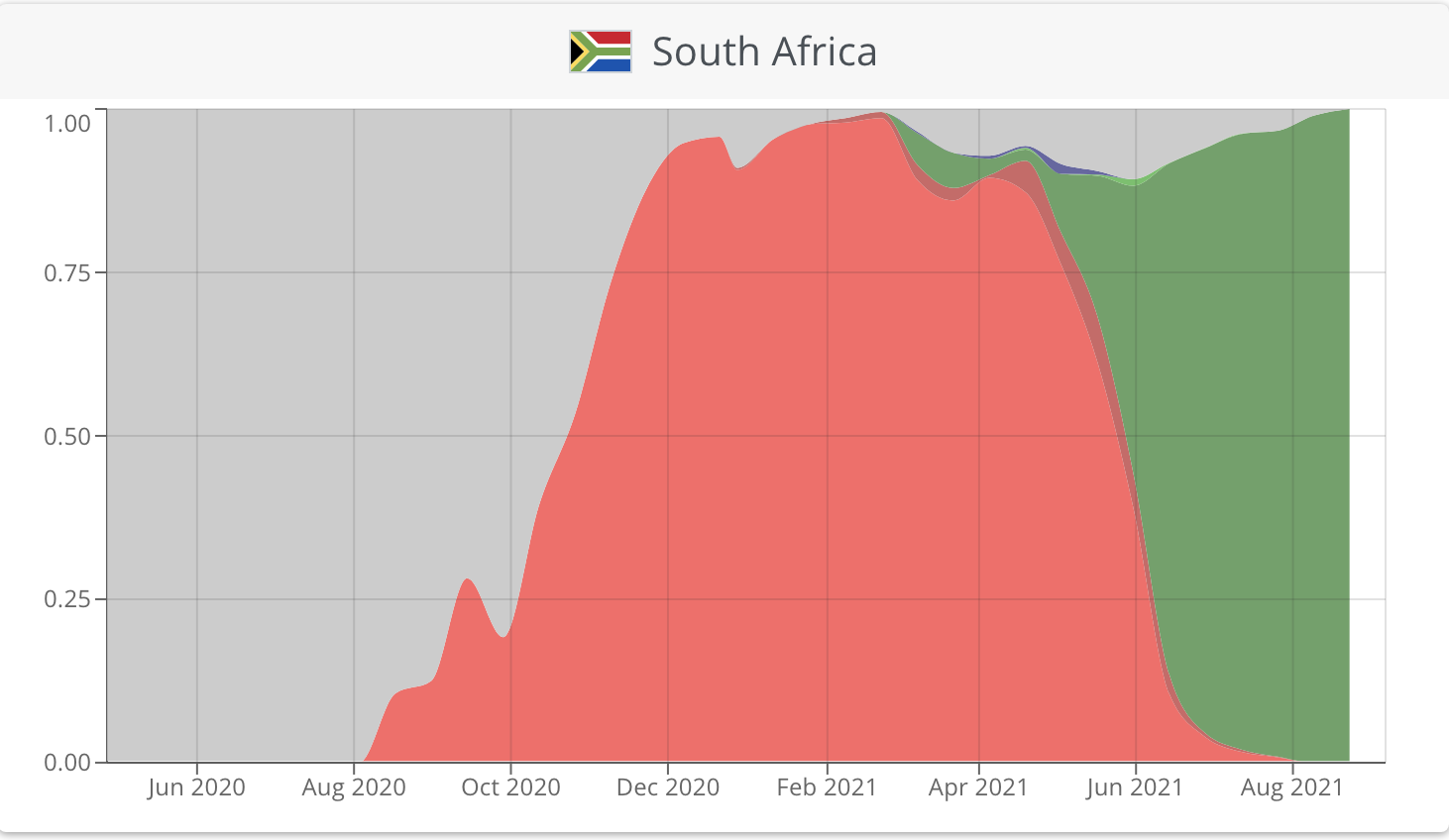
Figure 1: Plotting the prevalence of SARS-CoV-2 variants across time shows that the South African epidemic was initially dominated by the beta variant (red), but following its introduction, the delta variant (green) now dominates infections.
- To identify the emergence of new variants.
SARS-CoV-2 is continually evolving and, thus, it is critical to continue to monitor this evolution through sampling, sequencing and analysis of new infections across time. While surveillance may identify novel mutations or combinations of mutations within a sample of genomes, this does not necessarily mean that such variants pose an increased risk. The WHO has recommended a phased approach for designating variants. Variants with genetic changes that may pose a future risk but where there is no current evidence of phenotypic or epidemiological risk are added to a list for further monitoring. For example, the C.1.2 variant, first reported in South Africa in August 2021, has recently been added to this list. To date there is no evidence that C.1.2 has increased risks but variants that show such evidence can be flagged as variants of interest (VOIs) and if sufficient evidence exists, are eventually designated as variants of concern (VOCs).
To date, the Exatype platform has been used to characterize tens of thousands of genomes by laboratories all over the world.
In South Africa alone, over 15,000 genomes have been have been analysed by public sector institutions which is equivalent to 80% of South African SARS-CoV-2 genomes publicly available in GISAID. These outputs have been instrumental in monitoring and identifying SARS-CoV-2 variants across South Africa and have been used in a recent study tracking SARS-CoV-2 evolution across Africa1.
The unique features of Exatype enables laboratories to quickly and automatically perform SARS-CoV-2 variant typing on sequence data without any need for bioinformatics expertise or training:
- Ease of use – users drag and drop their raw sequence data onto their browser, click go, and Exatype does the rest.
- No training – Users need zero training in how to use the system and there’s no end-user input/time needed in processing the data.
- Speed - data is processed in parallel with results returned on average in ~1 hour. Other software used by a new Exatype user was taking up to 24 hours to process the data and produce results.
- Scalability - because Exatype is cloud-based, end-users don’t need to install/maintain hardware and Exatype autoscales meaning that there is zero restriction on the amount of samples that can be processed concurrently.
- Comprehensive outputs – Exatype outputs all the information needed for high-throughput genomic surveillance. For every sample, Exatype reports detected variants using both Pangolin lineages and Nextstrain clades nomenclature (always using the latest versions) and outputs can be directly uploaded to public resources (e.g. GISAID).
I’m so proud of the role that Exatype has played to date and, as countries across Africa and beyond continue to scale their SARS-CoV-2 genomic surveillance, we at Hyrax Biosciences are excited and prepared to continue to support the global response to the COVID-19 pandemic.

 Your sequence data.
Your sequence data.